Research in the Sander Lab
The goal of research in Dr. Sander’s laboratory is to understand the molecular mechanisms that underlie the formation and proper function of pancreatic insulin-producing beta cells, which are affected in diabetes. A major direction of the laboratory is to explore how dynamic changes in gene expression, transcription factor occupancy, and chromatin affect beta cell differentiation and maintenance of a functional beta cell state. To unravel these mechanisms, we combine next generation sequencing-based assays with molecular, cell-based, and genetic approaches in both mice and human pluripotent stem cells (hPSCs). Our work has uncovered fundamental mechanisms of cell fate determination and plasticity in the context of normal beta cell development, regeneration, and pathogenesis of diabetes.
Disease Modeling for Diabetes Using Pluripotent Stem Cells
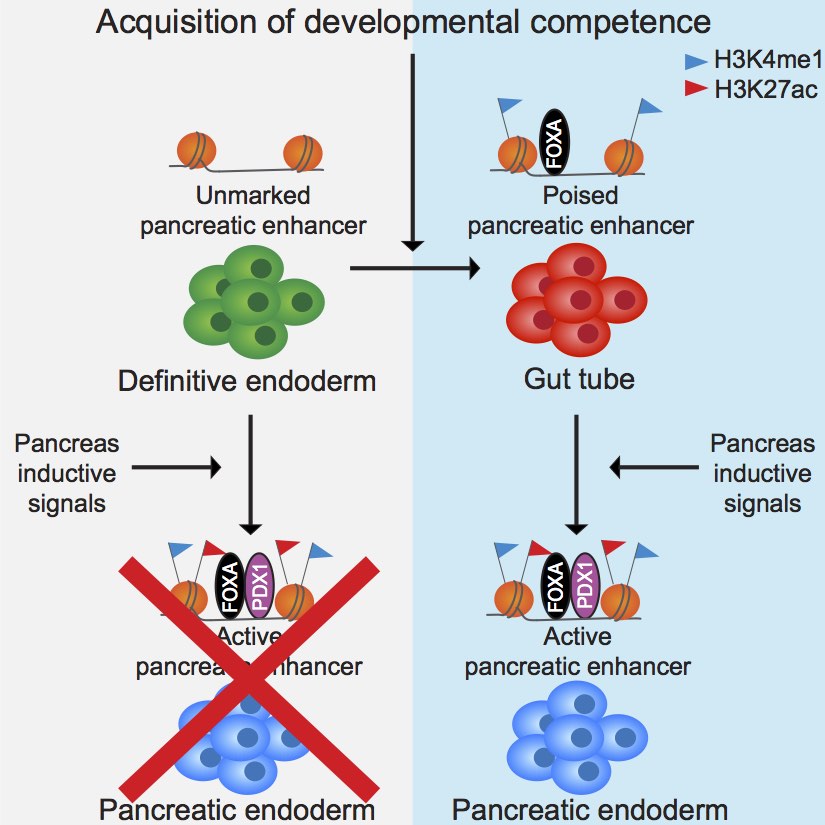 Diabetes is a group of complex diseases characterized by the inability to properly regulate blood glucose levels. Heredity plays an important role in determining who develops diabetes indicating a genetic basis for these diseases. Studies attempting to identify the genetic causes have revealed that the genetics associated with diabetes is complex and involves many different regions in the genome. Important unanswered questions include: How do genetic variants in isolation or combination cause diabetes? How can we identify target genes for variants that lie in non-coding regions of the genome?
Diabetes is a group of complex diseases characterized by the inability to properly regulate blood glucose levels. Heredity plays an important role in determining who develops diabetes indicating a genetic basis for these diseases. Studies attempting to identify the genetic causes have revealed that the genetics associated with diabetes is complex and involves many different regions in the genome. Important unanswered questions include: How do genetic variants in isolation or combination cause diabetes? How can we identify target genes for variants that lie in non-coding regions of the genome?
In a multi-disciplinary effort, our laboratory leverages human genetics, genomics, human stem cell engineering, and bioengineering approaches to address these and other important questions, focusing on the role of pancreatic beta cells in diabetes. Ongoing studies take advantage of our expertise in pancreas epigenomics to predict which diabetes-associated variants could be causal. By editing the genome of human pluripotent stem cells (hPSCs) via CRISPR-Cas9 and differentiating them using state-of-the-art pancreatic beta cell differentiation protocols, we aim to model the effects of specific variants and the role of their target genes in vitro. In collaboration with bioengineers, we are developing hPSC-based multi-cellular organoid models of the human islet to study how cell-cell interactions affect beta cells in health and disease. A specific focus is to understand the role of islet immune cells in the pathogenesis of type 1 and type 2 diabetes. The hope is that an improved understanding of gene regulatory pathways affecting beta cell function, regeneration, and survival will help identify novel treatments for individuals with diabetes. We are conducting this work as part of the NIH-funded T2D-GENES Consortium and Human Islet Research Network (HIRN).
Metabolic Regulation of Beta Cell Function
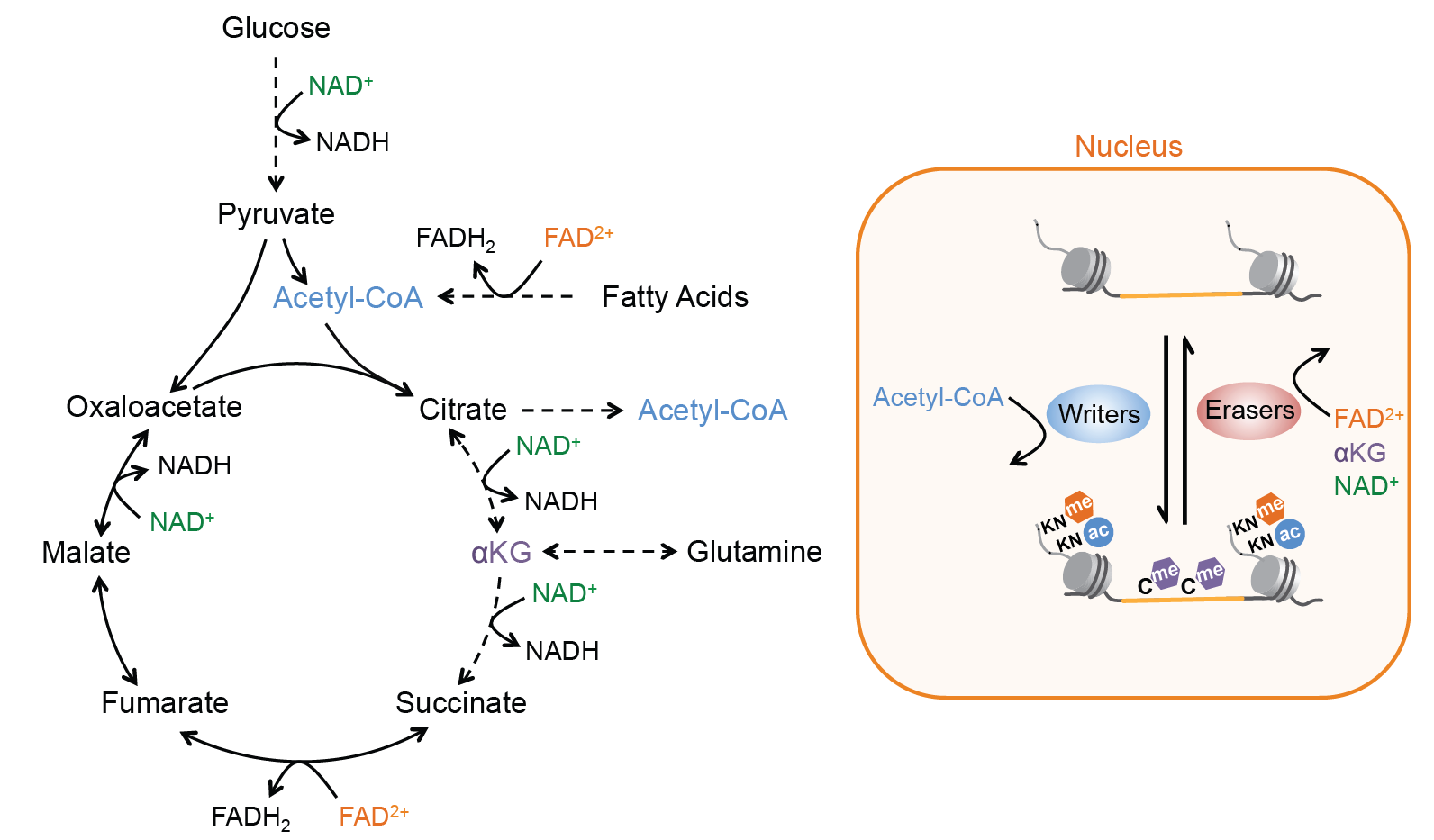 Pancreatic beta cells exhibit functional plasticity, such that past nutritional conditions influence future insulin secretory responses. In this way, insulin release is calibrated to anticipated insulin demand. Adaptability of beta cells enables tight control of blood glucose levels in the face of fluctuating metabolic conditions. Insufficient beta cell adaptation to changing nutrient states can result in diabetes or hypoglycemic disorders.
Pancreatic beta cells exhibit functional plasticity, such that past nutritional conditions influence future insulin secretory responses. In this way, insulin release is calibrated to anticipated insulin demand. Adaptability of beta cells enables tight control of blood glucose levels in the face of fluctuating metabolic conditions. Insufficient beta cell adaptation to changing nutrient states can result in diabetes or hypoglycemic disorders.
How does the beta cell “remember” prior metabolic conditions? Observations in other nutrient-sensitive cell types have implicated epigenetic modifications in cellular memory. Epigenetic marks, which include post-translational modifications of histones and DNA, are deposited by chromatin-modifying enzymes. These chromatin modifications dynamically respond to stimuli and can endure following removal of the stimulus, thereby influencing subsequent responses to the same signal. We propose that alterations to the epigenome could link past nutrient state to adaptive changes in beta cell function. Intriguingly, most chromatin-modifying enzymes utilize substrates or cofactors that fluctuate in response to metabolic conditions. We seek to identify the sensors and stimuli that link metabolic conditions to chromatin state, with the goal of identifying adaptive mechanisms in beta cells to respond to nutrient challenges. To this end, we apply a combination of genomic, metabolomic, mouse genetic, and islet physiological approaches to characterize the response of pancreatic beta cells to diverse nutritional challenges.
Beta Cell Maturation, Aging, Regeneration
 It is known that the regenerative capacity of beta cells declines rapidly with age. This decline is inversely correlated with an improved capacity of beta cells to regulate insulin secretion. Our group investigates the molecular pathways that are regulated during beta cell aging with the goal to identify therapeutic targets for expanding functional beta cell mass in patients with diabetes.
It is known that the regenerative capacity of beta cells declines rapidly with age. This decline is inversely correlated with an improved capacity of beta cells to regulate insulin secretion. Our group investigates the molecular pathways that are regulated during beta cell aging with the goal to identify therapeutic targets for expanding functional beta cell mass in patients with diabetes.
To define the molecular signatures associated with age-related changes in beta cells, we have employed innovative techniques, such as single-cell RNA sequencing and in vivo quantitative proteomics. These molecular profiling studies have revealed novel proteins and signaling pathways that regulate beta cell proliferation. Using rodent and human islets as a model, we are working to understand how beta cells integrate different signals to regulate their regenerative capacity and function. In collaboration with pharmaceutical companies, we are developing compounds to stimulate beta cell growth in humans.
